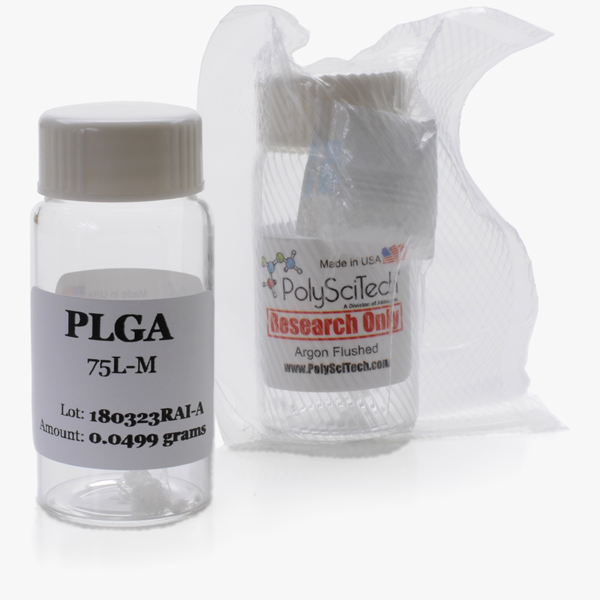
Standards
PolySciTech PLGA/PLA Standards are made by triple purifying each manufactured polymer:
- Dichloromethane filtration into non-solvent hexane
- Tetrahydrofuran filtration into non-solvent water
- Activated charcoal filtration from acetone
The purifying process is followed by high-temperature, deep-vacuum drying.
Each PLGA/PLA Standard comes as a convenient, pre-weighed aliquot of 50 mg (exact mass indicated on label) in an argon-flushed 20 ml vial and vacuum-packed with desiccant.
To use, add 10 ml of solvent to the vial to create a 5 mg/ml solution or add 20 ml of solvent to the vial to create a 2.5 mg/ml solution. These ready-to-use solutions can be applied for use in GPC type systems to calibrate to the PLGA itself.
The provided manufacturer data sheets for each standard includes state-of-the-art characterization:
- Gel-permeation with quadruple detection (refractive index, viscometer, multi-angle light scattering (17 different angles) and inline dynamic light scattering)
- Nuclear magnetic resonance (NMR)
- Gel-permeation correlation to commercial polystyrene standards
- Fourier-transform infrared spectrophotometry
GPC 4D
PLGA standards analyzed using GPC-TD. GPC-TD system consisted of an Agilent 1260 Infinity II HPLC connected to Dawn Heleos II (MALLS) coupled to Dynapro Nanostar DLS via optical cable, Optilab T-rEX (RI detector) and Viscostar III viscometer operated by Astra 7 software. GPC analysis performed by injecting 100 µl of ~2.0 mg/ml polymer solution. Separation performed with a linear gradient column (Tosoh Bioscience LLC, TSKgel GMHHR-L, 7.8 mm x 30 cm) at 0.6 ml/min flow of Acetone. Separation performed in acetone due to its greater dn/dc for PLGA as compared to tetrahydrofuran which is typically used for GPC analysis. In addition, individual dn/dc values experimentally determined for PLGA based on L:G ratio. This was performed utilizing batch method measurement in Astra 7 software using Optilab T-rEX. The obtained dn/dc values used for Astra 7 software calculations allow greater accuracy in light scattering data used for molar mass determination.
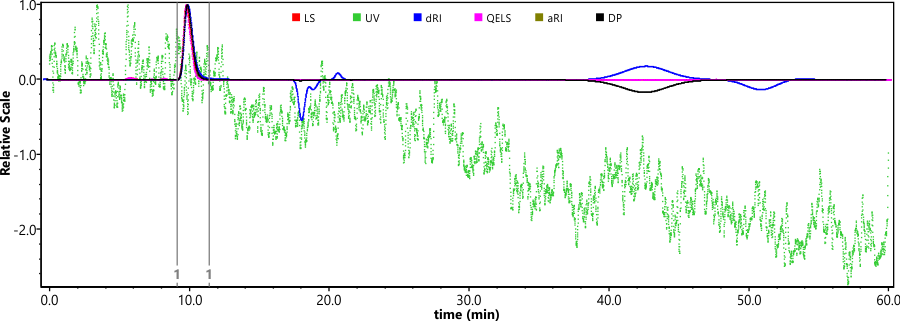
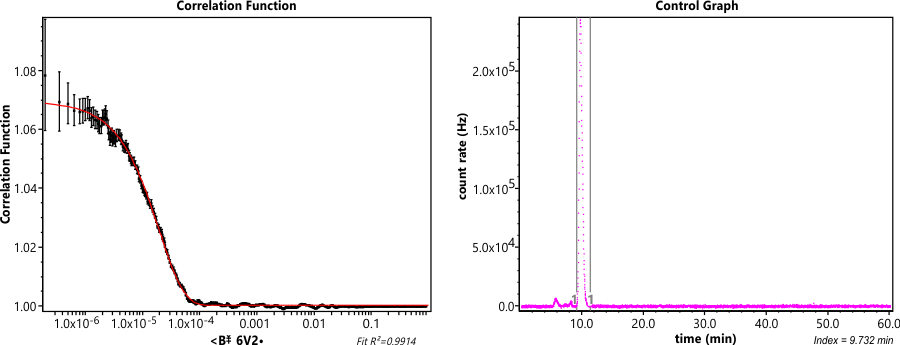
This assay gives the following information regarding the polymer:
| Value | Description |
|---|---|
| MHS Intercept (K) | Mark-Houwink constant “K” |
| MHS slope(a) | Mark-Houwink constant alpha |
| Mn (kDa) | Number average Molecular weight |
| Mp (kDa) | Peak molecular weight |
| Mv (kDa) | Viscosity average molecular weight |
| Mw (kDa) | Weight average molecular weight |
| Mz (kDa) | Z-average molecular weight |
| Polydispersity (Mw/Mn) | Distribution of molecular mass |
| rn (nm) | Number-average mean square radius |
| rw (nm) | Weight-average mean square radius |
| rz (nm) | Z average radius |
| r(avg) (nm) | Average mean square radius |
| rh(v)n (nm) | Number-average hydrodynamic radius |
| rh(v)w (nm) | Weight-avg mean hydrodynamic radius |
| rh(v)z (nm) | Z-average hydrodynamic radius |
| rh(v)(avg) | Average hydrodynamic radius |
| [η]n (mL/g) | Number-average intrinsic viscosity |
| [η]w (mL/g) | Weight-average intrinsic viscosity |
| [η]z (mL/g) | Z-average intrinsic viscosity |
| dn/dc | Refractive index increment |
NMR
Samples are prepared as 5-10 mg/ml solutions in deuterated chloroform and spectroscopy is collected at Purdue Interdepartmental NMR Facility. The collected spectra are shown on the data-file along with a few calculations that are performed:
Lactide:Glycolide Ratio
The collected spectra are analyzed to obtain LA:GA ratio by proton counting. The exact method is described in a previous Akina publication but briefly, the intensities at 5.2 ppm and 4.8 ppm are used for this. The integration at 5.2 ppm (PL) is ascribed to a single H of lactide while the integration at 4.8 ppm (PG) is ascribed to 2H of glycolide. The molar ratio of lactide ML in the PLGA is calculated as follows:
ML = PL/(PL+(PG/2))
The molar ratio of glycolide is 100% - ML.
The weight ratio of LA:GA is calculated by multiplying the molar ratio of each component by its monomeric residue molecular weight which is 58.04 g/mol for glycolate and 72.06 g/mol for lactate, respectively.
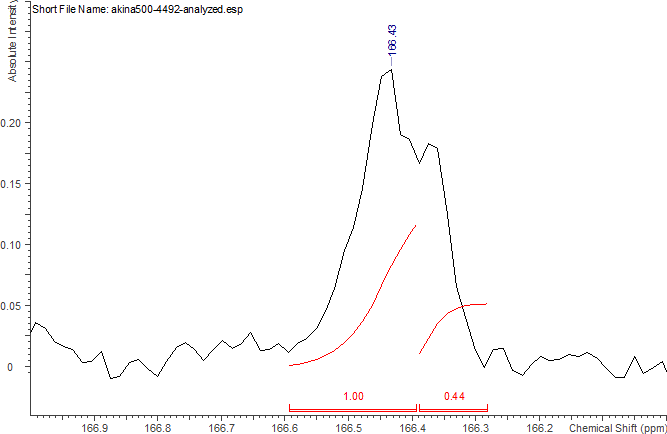
Blockiness (Rcms)
Although PLGA’s are made to be random copolymers, the lower melting point and preferential reaction rate for glycolide causes the creation of regions of higher glycolide content and subsequently regions of higher lactide content. The degree and incidence of these regions of relatively higher content of one monomer over the other is referred to as ‘blockiness’ and is measured by Rcms which is the ratio of glycolides next to lactides (random regions) to glycolides next to other glycolides (block regions).
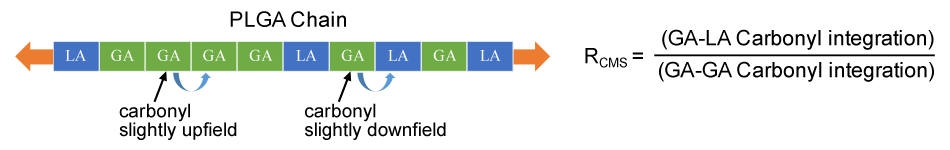
Determination of Rcms (blockiness) in a PLGA gives information about the relative position of lactides and glycolides along the chain is done for these standards using C13NMR spectroscopy according to this publication. For this, the C13NMR peaks for glycolide carbonyl carbon at 166 - 167 ppm are compared. The integration of the downfield carbonyl peak (GA next to LA) is divided by the upfield carbonyl peak (GA next to GA).
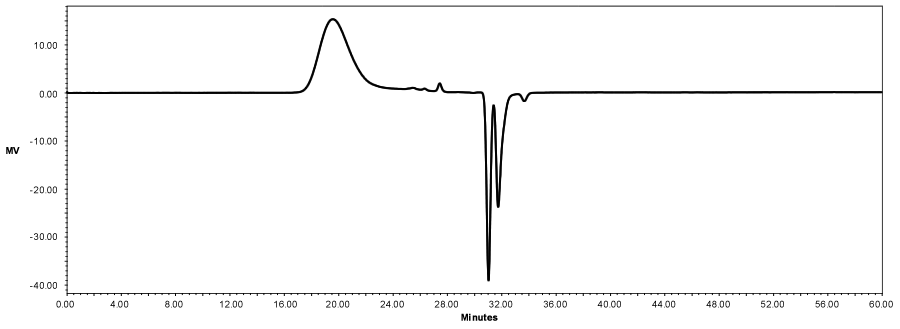
GPC External Standards
PLGA standards analyzed using GPC. GPC system consisted of Waters 1515 Isocratic HPLC pump connected to Waters 2707 Autosampler and Waters 2414 Refractive Index Detector. GPC analysis preformed by injecting 100µL of ~ 2.0 mg per mL polymer solution dissolved in 2.0 µm filtered THF. A run time of 60 minutes was set with the flow rate of 1mL THF/min, and separation performed by a series of three GPC columns. The first one the samples passed through is a Phenomenex column Phenogel 5µ 50A 300x7.5mm, the second is Phenomenex column Phenogel 5µ 10E4A 300x7.5, and the last one is Aglient Resipore 300x7.5mm 3µm column. These samples were tested against Agilent Technologies EasiCal PS2 polystyrene standards lot numbers PL2010-0601, PL2010-0605. These standards were prepared according to instructions using 0.2 µm filtered THF.
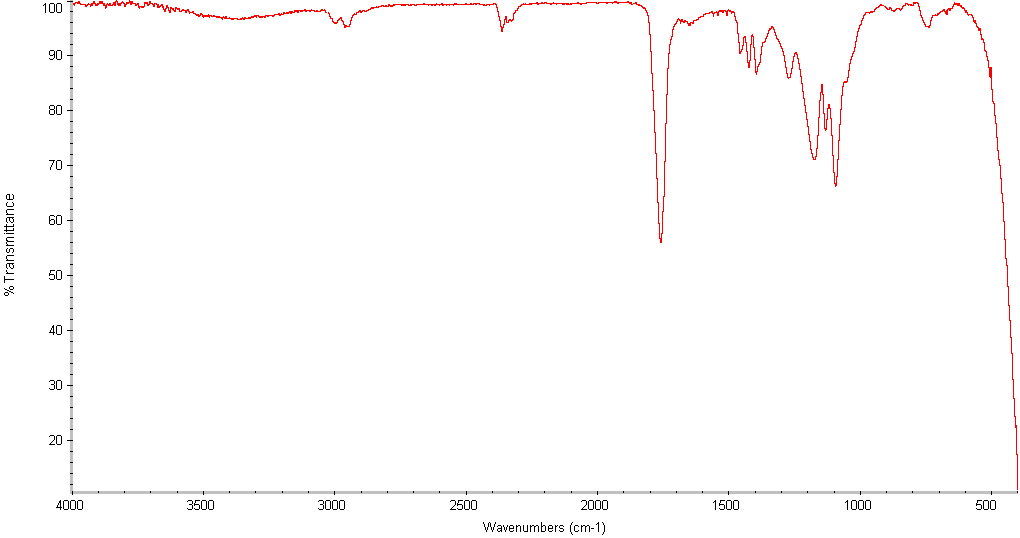
FTIR
PLGA standards analyzed using FTIR. FTIR system consisted of Themo Nicolet Avatar 329 FT-IT. Samples were prepared by dissolving polymer in THF at a concentration of 2mg/mL, then placed on KBr pellets and deep vacuum dried to remove all solvent before analysis.
 Akinalytics
Akinalytics Midwest GMP
Midwest GMP Polymer Blog
Polymer Blog








